Degpred: Systematic Prediction of Degrons and Binding E3 ligases via Deep Learning
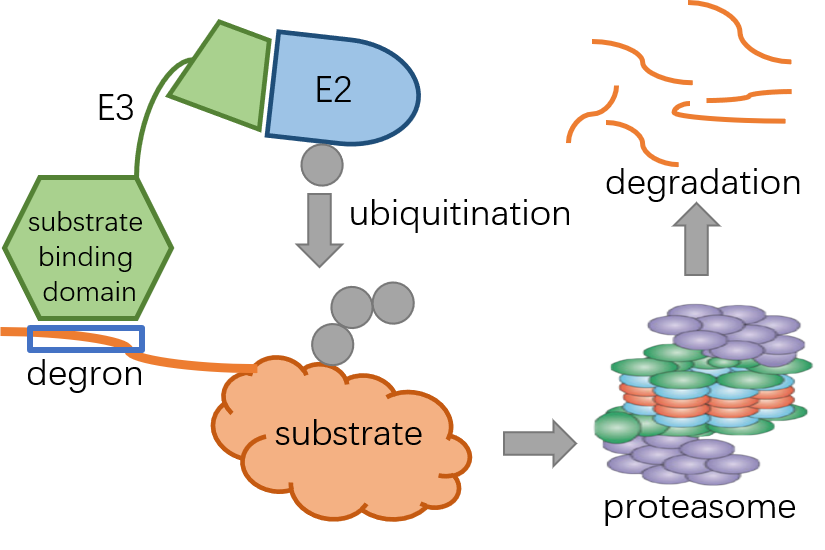
Degrons are short linear motifs, bound by E3 ubiquitin ligases to target proteins to be degraded by the ubiquitin-proteasome system. Deregulation of degron disrupts control of protein abundance and commonly contributes to diseases. Despite with important functions, only a limited number of degrons have been identified by experiment, the widely used motif matching prediction method is limited by few motifs and high false positive rate. Here, we developed a deep learning model Degpred to predict degrons directly from protein sequences. Leveraging abundant protein features provided by the BERT based model, Degpred predicts degrons beyond those from known motifs and greatly expands the degron landscape. Degpred outperformed motif-based methods in capturing well-known degron properties. Furthermore, we calculated motifs for 39 E3s using our collected E3-substrate interaction dataset and assigned predicted degrons to specific E3s. In summary, we presented an efficient and general tool to predict degrons and binding E3s, both collected and predicted datasets were integrated in this website.
News
2022-05-11: We added known degrons and E3-substrate interactions, and the protein function annotations.
2021-12-13: Degpred results for UniProt human reviewed proteins were provided.
Chao Hou, Yuxuan Li, Mengyao Wang, Hong Wu, Tingting Li. Systematic Prediction of Degrons and Binding E3 ligases via Deep Learning